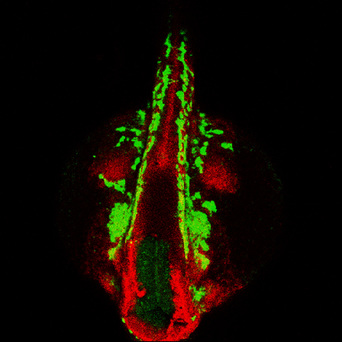
Genetic and epigenetic regulation of neural crest development
What are the gene regulatory networks involved in the specification and differentiation of neural crest cells? Here, we are focusing on the transcriptional regulation of these processes by the zinc finger transcription factor family of Prdm proteins. When prdm1a is mutated in zebrafish, neural crest and Rohon-Beard sensory neurons are reduced, and the neural plate domain is expanded. Thus, Prdm1 controls the cell fate decision between neural crest cells and other cells at the neural plate border by both transcriptionally repressing and activating transcriptional targets. In the craniofacial region, prdm1a and prdm3 and prdm16 proteins play important roles in craniofacial development by regulating the downstream Wnt signaling pathway. Currently, we are determining how Prdm1, Prdm3, and Prdm16 function as epigenetic regulators and interact with other transcriptional regulators and chromatin modifiers and signals to regulate neural crest development and differentiation into cartilage as well as cranial ganglia including the trigeminal ganglion.
What are the gene regulatory networks involved in the specification and differentiation of neural crest cells? Here, we are focusing on the transcriptional regulation of these processes by the zinc finger transcription factor family of Prdm proteins. When prdm1a is mutated in zebrafish, neural crest and Rohon-Beard sensory neurons are reduced, and the neural plate domain is expanded. Thus, Prdm1 controls the cell fate decision between neural crest cells and other cells at the neural plate border by both transcriptionally repressing and activating transcriptional targets. In the craniofacial region, prdm1a and prdm3 and prdm16 proteins play important roles in craniofacial development by regulating the downstream Wnt signaling pathway. Currently, we are determining how Prdm1, Prdm3, and Prdm16 function as epigenetic regulators and interact with other transcriptional regulators and chromatin modifiers and signals to regulate neural crest development and differentiation into cartilage as well as cranial ganglia including the trigeminal ganglion.
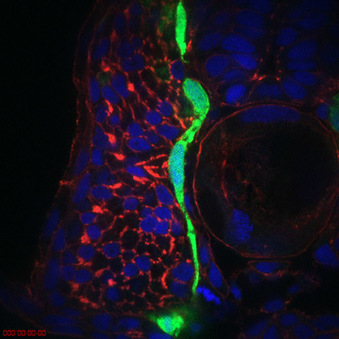
Cellular mechanisms of neural crest migration
How do neural crest cells migrate along the correct migratory pathway? Transcriptional profiling suggests that many of the genes regulated by neural plate border transcription factors are involved in cell migration. We are testing the hypothesis that neural plate border transcription factors regulate the processes of neural crest cell migration including genes such as chemokine signaling, cell-cell adhesion and cell-matrix adhesion genes. One such factor, cxcr4a, is expressed on cranial neural crest cells and sdf1b is expressed in the target tissue. Loss of cxcr4a causes neural crest migration and craniofacial cartilage and ganglia condensation defects. Current work is focused on understanding the molecular mechanisms as well as identifying several other downstream targets involved in this process including boc and cdon.
How do neural crest cells migrate along the correct migratory pathway? Transcriptional profiling suggests that many of the genes regulated by neural plate border transcription factors are involved in cell migration. We are testing the hypothesis that neural plate border transcription factors regulate the processes of neural crest cell migration including genes such as chemokine signaling, cell-cell adhesion and cell-matrix adhesion genes. One such factor, cxcr4a, is expressed on cranial neural crest cells and sdf1b is expressed in the target tissue. Loss of cxcr4a causes neural crest migration and craniofacial cartilage and ganglia condensation defects. Current work is focused on understanding the molecular mechanisms as well as identifying several other downstream targets involved in this process including boc and cdon.
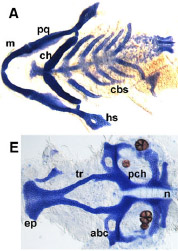
Understanding Craniofacial disease
How does the craniofacial skeleton form during development? Many complex morphogenetic processes rely on genetic and epigenetic information to form a proper face. Interestingly, the PRDM family of transcription factors have been correlated with human craniofacial disease such as cleft lip and palate, and other transcriptional regulators we study are linked to syndromes such as Split Hand Foot Malformation and Treacher-Collins Syndrome. We are interested in how these genes that are involved in development are mutated in disease. Currently, we are determining how Prdm1 functions as a transcriptional regulator to pattern the vertebrate face and limbs, as well as prdm3/prdm16 in cleft lip and palate as well as TMD.
How does the craniofacial skeleton form during development? Many complex morphogenetic processes rely on genetic and epigenetic information to form a proper face. Interestingly, the PRDM family of transcription factors have been correlated with human craniofacial disease such as cleft lip and palate, and other transcriptional regulators we study are linked to syndromes such as Split Hand Foot Malformation and Treacher-Collins Syndrome. We are interested in how these genes that are involved in development are mutated in disease. Currently, we are determining how Prdm1 functions as a transcriptional regulator to pattern the vertebrate face and limbs, as well as prdm3/prdm16 in cleft lip and palate as well as TMD.
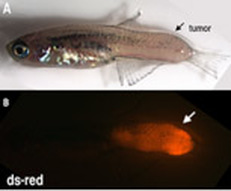
The role of developmental genes in cancer
Many developmental genetic regulators, including Six1 and Prdm1 among others are reactivated or highly mutated in human cancers. We have built zebrafish models of cancer, including rhabdomyosarcoma, MPNST and melanoma to determine the genetic and epigenetic regulation of our favorite developmental genes and their targets in cancer.
Many developmental genetic regulators, including Six1 and Prdm1 among others are reactivated or highly mutated in human cancers. We have built zebrafish models of cancer, including rhabdomyosarcoma, MPNST and melanoma to determine the genetic and epigenetic regulation of our favorite developmental genes and their targets in cancer.